Abstract
Rapid nucleic acid testing is central to infectious disease surveillance. Here, we report an assay for rapid COVID-19 testing and its implementation in a prototype microfluidic device. The assay, which we named DISCoVER (for diagnostics with coronavirus enzymatic reporting), involves extraction-free sample lysis via shelf-stable and low-cost reagents, multiplexed isothermal RNA amplification followed by T7 transcription, and Cas13-mediated cleavage of a quenched fluorophore. The device consists of a single-use gravity-driven microfluidic cartridge inserted into a compact instrument for automated running of the assay and readout of fluorescence within 60 min. DISCoVER can detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in saliva with a sensitivity of 40 copies μl–1, and was 94% sensitive and 100% specific when validated (against quantitative PCR) using total RNA extracted from 63 nasal-swab samples (33 SARS-CoV-2-positive, with cycle-threshold values of 13–35). The device correctly identified all tested clinical saliva samples (10 SARS-CoV-2-positive out of 13, with cycle-threshold values of 23–31). Rapid point-of-care nucleic acid testing may broaden the use of molecular diagnostics.
Similar content being viewed by others
Main
Rapid, point-of-care nucleic acid detection is a critical component of a robust testing infrastructure for controlling disease transmission. Outbreaks such as the coronavirus disease 2019 (COVID-19) pandemic have highlighted limitations of the centralized diagnostic laboratory model and its lengthy sample-to-answer time. Laboratory-developed tests such as quantitative polymerase chain reaction (qPCR) are conducted in facilities that require labour-intensive personnel and equipment infrastructure for sample accessioning, nucleic acid extraction, thermocycling and data analysis. Sample transport and result reporting time also greatly contribute to the turnaround times of centralized tests. The development of an easy-to-use microfluidic device paired with on-site detection would enable increased volume of and access to rapid molecular testing.
So far, well over 6 million deaths from COVID-19 have resulted from over 500 million infections by its causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The myriad challenges of a high asymptomatic infection rate1, insufficient testing and the narrow time window for molecular tests to be highly sensitive2,3 can be combatted by broad deployment of on-site molecular diagnostics, such as upon entry into the workplace or classroom. There is also tremendous potential for community surveillance testing to augment clinical workflows, where positive cases are confirmed by referral to a more constrained supply of clinical-grade tests. Alternative sampling methods and test technologies can also help diversify the diagnostic supply chain, as the standard pipeline for clinical testing can be limited by RNA extraction kit or swab shortages4,5.
We therefore sought to develop a method that did not require RNA extraction or upper respiratory tract swabs and could be integrated into a rapid, automated microfluidic workflow. qPCR-based assays have previously been developed with direct sample spike-in, and typically use a high temperature for sample lysis6. Other approaches have exploited chaotropic agents, chemical reduction and RNase inhibitors6,7,8,9. Furthermore, saliva samples are reported to have 97% concordance with nasopharyngeal (NP) swabs10,11.
Clustered regularly interspaced short palindromic repeats (CRISPR)-based detection is a promising new approach for nucleic acid diagnostics. These methods rely on the guide-RNA-dependent activation of Cas13 or Cas12 nucleases to induce non-specific single-stranded RNA or single-stranded DNA nuclease activity, respectively, in order to cleave and release a caged reporter molecule12,13,14,15,16,17,18,19. The released reporter can be quantitatively measured with a fluorescence detector to read out the test result. CRISPR-based detection is highly specific, but Cas13 nucleases alone can take up to 2 h to reach attomolar sensitivity for diagnostic applications20,21. In contrast, loop-mediated isothermal amplification (LAMP) performs highly sensitive nucleic acid amplification in under 20 min with attomolar limits of detection20 (LODs). However, despite the sensitivity and speed of LAMP, such isothermal methods are often prone to non-specific amplification22,23.
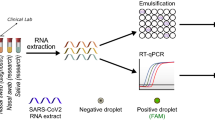
In this Article, to address these challenges, we report DISCoVER (for diagnostics with coronavirus enzymatic reporting), an RNA-extraction-free test that combines two distinct amplification mechanisms for sensitivity with a Cas13-mediated probe for specificity (Fig. 1). Following direct lysis and inactivation of saliva samples via denaturation and reduction, LAMP amplification is followed by T7 transcription to provide two layers of target amplification. To demonstrate viral CRISPR testing in a sample-to-answer format, we integrated the DISCoVER workflow into a microfluidic system with a compact fluorescence reader for real-time detection. In a point-of-care proof of concept, sample inactivation and subsequent amplification and detection with this device can be stacked such that the inactivation time is not rate-limiting in the workflow (Supplementary Fig. 1).
Patient samples, such as saliva, are collected and heat-inactivated in direct lysis buffer, followed by loading onto a single-use, gravity-driven microfluidic cartridge. The inactivation step ranges from 5 to 20 min depending on institutional regulations37. The cartridge is then inserted into a companion instrument that automatically runs the DISCoVER assay in a closed system to minimize reaction contamination. In step 1, an initial rLAMP reaction employs two mechanisms for amplification of target nucleic acids. RFU, relative fluorescence units. Cas13 enzymes are programmed with a guide RNA to specifically recognize the desired RNA molecules over non-specifically amplified products. Subsequent activation of Cas13 ribonuclease activity, in step 2, results in cleavage of reporter molecules for saturated signals within 5 min of CRISPR detection. Including actuation time of the device, time to readout in a finalized system is 48 min. In the left half of the cartridge, guide RNAs targeting SARS-CoV-2 enable rapid and selective detection of attomolar concentrations of virus. The mirrored half of the cartridge is used for an internal process control, enabling a negative test result by ensuring the presence of adequate patient samples. By exploiting template switching and CRISPR programmability, the point-of-care DISCoVER system can contribute to increased surveillance of diverse pathogens.
DISCoVER incorporates a 30-min amplification step followed by a 5-min Cas13 readout, and achieves attomolar sensitivity on unextracted saliva samples. We also multiplex the amplification with a process control based on detection of a reference human gene to confirm the presence of sufficient cellular material in the sample. We validated DISCoVER on a saliva-based sample matrix containing live SARS-CoV-2 virus and total RNA extracted from patient nasal swabs with cycle threshold (Ct) values ranging from 15 to 35. We observed a positive predictive value (PPV) of 100% and a negative predictive value (NPV) of 93.75% for the DISCoVER assay relative to qPCR. Contrived positive saliva samples were assayed end-to-end on a microfluidic device with integrated real-time fluorescence detection as proof of concept for automated detection. An on-board process control was used to allow for the discrimination between positive, negative and invalid test results. To further validate the robustness of the automated microfluidic system, contrived saliva samples were run, and indicated attomolar sensitivity. Finally, field-collected, natural clinical samples were run on the device to demonstrate the assay’s capacity to differentiate positive and negative real-patient samples. Overall, the DISCoVER system enables portable, rapid and sensitive viral detection for pathogen surveillance.
Results
We first sought to compare the nucleic acid detection properties of Cas13 and Cas12 enzymes, assessing the reporter cleavage activity of Leptotrichia buccalis Cas13a (LbuCas13a) and Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a) with matching guide RNA spacer sequences in a dilution series of their corresponding synthetic activator molecules (Fig. 2a). We observed that LbuCas13a detection is substantially faster than LbCas12a at low activator concentrations. At 100 pM of this activator sequence, LbuCas13a reaches half-maximum fluorescence over 30 times faster than LbCas12a in their respective reaction conditions (Fig. 2b and Methods). However, the LOD of LbuCas13a is still in the femtomolar range after 60 min (ref. 24), which is already beyond the maximum sample-to-answer time that is likely to be relevant for a point-of-care assay.
a, Cas13 and Cas12 detection kinetics at varying activator concentrations. Line and shaded regions denote mean ± standard deviation (s.d.) with n = 3 technical replicates. b, Cas13 and Cas12 time to half-maximum fluorescence. †, time to half-maximum fluorescence was too rapid for reliable detection. ††, time to half-maximum fluorescence could not be determined within the 120 min assay runtime. Values are mean ± s.d. with n = 2 technical replicates. c, Schematic of SARS-CoV-2 genome sequence, with LAMP primer set locations indicated. d, Representative fluorescence plots of LAMP amplification of 100 copies μl–1 of synthetic SARS-CoV-2 RNA or NTC. Shaded regions denote mean ± s.d. with n = 3 technical replicates. e, Time to threshold of nine screened LAMP primer sets, targeting synthetic SARS-CoV-2 RNA or NTC. Replicates that did not amplify are represented at 0 min. Error bars represent mean ± s.d. of amplified technical replicates, n = 4. f, LOD assay of LAMP using N set 1 primer set. Replicates that did not amplify are represented at 0 min. Error bars represent mean ± s.d. of amplified technical replicates, n = 4.
To achieve attomolar sensitivity, we chose LAMP as a cost-effective and rapid method for isothermal amplification. LAMP employs a reverse transcriptase, a strand-displacing DNA polymerase, and three sets of primer pairs to convert viral RNA to DNA substrates for LAMP. We screened nine LAMP primer sets targeting distinct regions across the length of the SARS-CoV-2 genome (Fig. 2c and Supplementary Table 1)25,26,27,28. When targeted to SARS-CoV-2 genomic RNA fragments at 100 copies μl–1, all sets resulted in positive LAMP signals. Maximum fluorescence was reached within 20 min for all primer sets, and time to threshold was determined via the single-threshold cycle quantification (Cq) determination mode as indicated (Fig. 2d). LAMP primer sets targeting Orf1ab set 1, N set 1 and N set 2 consistently amplified in under 15 min (Fig. 2e).
Each LAMP primer set resulted in a no-template control (NTC) signal, albeit with a delay relative to the positive condition containing viral RNA (Fig. 2d). This high false-positive rate, potentially due to primer dimer formation, can in principle be reduced with a second probe that selectively recognizes the amplified nucleic acid sequence. We therefore sought to combine the sensitivity of LAMP detection with the specificity of Cas13 target recognition.
To avoid detection of non-specific amplification products by Cas13, the guide RNAs must have minimal sequence overlap with the primer sequences. Owing to the complexity of LAMP concatemerization, LAMP primers are highly overlapping and amplify short target regions to increase the reaction speed24,29. Our LAMP primer sets generated amplicons with primer non-overlapping sequences ranging from 1 to 60 nt. N set 1, targeting the SARS-CoV-2 N gene, was chosen for further use owing to its low time to threshold and an amplicon size capable of accommodating Cas13 guide RNAs (Fig. 2e and Supplementary Table 1). We next performed a dilution series of genomic viral RNA and determined the N set 1 LOD to be 25 copies μl–1 (Fig. 2f), comparable to previous studies that report LODs between 10 and 100 copies μl–1 (refs. 7,30,31).
As Cas13 targets single-stranded RNA (Fig. 2a), while LAMP amplifies DNA substrates, we employed transcription of the LAMP products to enable substrate compatibility. T7 RNA polymerase promoter sequences were incorporated into the LAMP primer sequences for subsequent transcription and Cas13 detection. We termed this ‘conversion of LAMP amplification to RNA’ or ‘rLAMP’. LAMP employs three primer pairs: forward and backward outer primers (F3/B3) for initial target strand displacement, forward and backward inner primers (FIP/BIP) to form the core LAMP stem–loop structure, and forward and reverse loop primers (FLoop/BLoop) for an additional layer of loop-based amplification (Extended Data Fig. 1). Through multiple iterations of primer binding and extension, these stem–loop structures amplify into concatemers composed of inverted repeats of the target sequence (Fig. 3a).
a, Schematic of rLAMP mechanism for exponential DNA amplification using F3/B3 and FIP/BIP primers, resulting in higher-order inverted repeat structures. Red arrows indicate location of T7 promoter sequence, inserted in the mBIP primer. Upon T7 transcription, the resulting RNA contains one or more copies of the Cas13 target sequence (orange). b, Schematic of the location of different T7 promoter locations on the rLAMP dumbbell structure and loop primers. c, rLAMP time to threshold of six distinct T7 promoter insertions. Replicates that did not amplify are represented at 0 min. Error bars represent mean ± s.d. of amplified technical replicates, n = 4. d, Denaturing PAGE gels of mBIP rLAMP products to verify T7-mediated transcription. AfeI cleaves in the crRNA target region of templated products, which is expected to result in a single major transcribed species. e, Kinetics of T7 transcription and Cas13 detection on mBIP rLAMP products. Values are mean ± s.d. with n = 3 technical replicates. f, Cas13 detection of eight technical replicates of mBIP rLAMP amplification on genomic RNA, depicted as fold change over NTC at different reaction endpoints.
To enable RNA transcription following LAMP (rLAMP), we systematically tested the insertion of T7 promoter sequences in three different regions of LAMP primers: on the 5′ end of the FIP and BIP primers (5′ FIP/5′ BIP), in the middle of the FIP and BIP primers (mFIP/mBIP), and on the 5′ end of the loop primers (FLoop/BLoop) (Fig. 3b). Addition of the T7 promoter did not greatly affect rLAMP time to threshold of N set 1, given viral genomic RNA at 100 copies μl–1 (Fig. 3c). To confirm the target sequences were specifically amplified, we performed restriction enzyme digestion on the LAMP products using AfeI, which digests in the Cas13 guide RNA target region within the rLAMP amplicon (Supplementary Fig. 2a,b). Lack of AfeI digestion in all NTC conditions confirmed that the NTC signal results from non-specific amplification of sequences lacking the guide-matching target sequence.
To test whether the T7 promoter was properly incorporated and functional in the rLAMP amplicon, we next performed in vitro transcription with T7 RNA polymerase. Denaturing polyacrylamide gel electrophoresis (PAGE) analysis indicated that the viral template and NTC conditions resulted in RNA transcription for all primer sets (Fig. 3d). As expected, AfeI digestion of the mBIP rLAMP product produced a single 147-nt product containing the T7 promoter (Supplementary Fig. 2a). Subsequent T7 transcription resulted in the expected 85-nt RNA product (Fig. 3d).
Next, we optimized reaction buffer conditions to support T7 transcription and Cas13 detection in a single step, followed by systematic screening of Cas13 cleavage activity in the presence of rLAMP products containing T7 promoter insertions in different rLAMP amplicons. We determined that the middle of the BIP primer (mBIP) insertion position resulted in the fastest detection and therefore chose this primer set for further studies (Supplementary Fig. 3 and Supplementary Table 2). Cas13 rapidly detects the rLAMP amplicon with a viral template, while avoiding detection of non-specific NTC amplicons, achieving over ten-fold change in signal over the NTC background in under 2 min (Fig. 3e). Signal saturation occurred within 5 min, reaching a signal-to-background ratio of over 40.
To confirm replicability of the DISCoVER pipeline, we tested Cas13 detection on eight replicates of mBIP rLAMP reactions with activator RNA included at a concentration of 100 copies μl–1 in a 20 µl reaction. All eight replicates resulted in a >25-fold increase in signal over NTC, which remains stable well beyond the 5-min detection time employed here (Fig. 3f).
With an amplification and detection protocol in place, we next optimized sample processing to establish a simple protocol of heat paired with chemical reagents to promote viral inactivation and dampen the activity of RNA-degrading nucleases present in saliva32. We assayed two concentrations of the shelf-stable reducing agent Tris(2-carboxyethyl)phosphine (TCEP) paired with the ion chelator ethylenediaminetetraacetic acid (EDTA) (refs. 7,8), commercially available reagents such as QuickExtract buffers containing detergents and proteinases, and DNA/RNA Shield containing chaotropic guanidine thiocyanate.
To test the compatibility of these inactivating reagents with rLAMP, we created mock-positive saliva samples by adding the reagents to heat-treated saliva at 75 °C (ref. 33) and adding SARS-CoV-2 genomic RNA at two different concentrations: 1,000 copies µl−1 and 200 copies µl−1. In the absence of inactivating reagents, we were unable to detect any rLAMP signal, suggesting degradation of RNA in saliva by endogenous RNases present in the sample (Fig. 4a). In contrast, genomic RNA without saliva was rapidly amplified in under 15 min. Only the low-concentration condition of TCEP–EDTA protected target RNA and preserved rLAMP sensitivity in all four replicates (Fig. 4a). This reagent cocktail simultaneously breaks protein disulfide bonds and sequesters divalent cations. Its activity is expected to dampen RNase activity while simultaneously disrupting mucin gel formation, reducing variable saliva viscosity for simpler sample processing34,35.
a, Direct saliva lysis conditions were tested for compatibility with the DISCoVER workflow. Replicates that did not amplify are represented at 0 min. Error bars represent mean ± s.d. of amplified technical replicates, n = 4. IA, inactivation reagent; QE, QuickExtract. b, Schematic of contrived saliva sample generation, quantification via RT–qPCR, and detection via DISCoVER to determine analytical sensitivity. c, Fold change in DISCoVER signal relative to NTC on SARS-CoV-2-positive saliva samples at 5 min of Cas13 detection. Error bars show mean ± s.d. with n = 20 biological replicates. d, Fold change in DISCoVER signal relative to NTC on 30 negative saliva samples collected before November 2019, at 5 min of Cas13 detection.
Next, we sought to determine the analytical sensitivity and specificity for DISCoVER on saliva samples (Food and Drug Administration, 2020). To determine the LOD, viral stocks were serially diluted in medium and quantified with qPCR with reverse transcription (RT–qPCR) relative to a standard curve generated from synthetic genomic RNA. These known concentrations of virus were spiked into negative saliva samples collected before November 2019 in BSL3 conditions and run through the DISCoVER workflow (Fig. 4b). We performed 20 DISCoVER replicates for a range of virus concentrations, determining 40 copies µl−1 of virus in directly lysed saliva (Fig. 4c) to be the lowest concentration tested where at least 19/20 replicates were amplified. To assay DISCoVER specificity, saliva samples from 30 different individuals negative for SARS-CoV-2 were tested without any false positive signal (Fig. 4d). The addition of influenza A H1N1 and H3N2, influenza B and human coronavirus OC43 synthetic RNA did not inhibit the detection of SARS-CoV-2, nor result in non-specific detection, further validating the specificity of this assay (Supplementary Fig. 4a). Additionally, we validated DISCoVER on multiple SARS-CoV-2 variants of concern, including the highly transmissible B.1.1.7 variant36 (Supplementary Fig. 4b).
In point-of-care settings, sample inactivation and lysis would ideally be confirmed by an internal process control (Fig. 5a). This is particularly important for saliva-based population screening, as some individuals can have high viscosity or mucin gel loads in their samples. According to Food and Drug Administration guidelines, a positive internal process control is necessary to determine if a sample is SARS-CoV-2 negative, while indicating proper sample collection, nucleic acid extraction, assay set-up and reagent functioning (‘CDC 2019-nCoV Real-Time RT-PCR Diagnostic Panel - Instructions for Use’; https://www.fda.gov/media/134922/download). If the internal process control is negative in a clinical specimen, the result should be considered invalid, unless SARS-CoV-2 is detected, in which case the sample is presumed positive. To achieve this, we multiplexed amplification by adding both N gene and human RNase P gene primers into the rLAMP step. Subsequent Cas13 detection of RNase P on saliva samples reached saturation within 5 min (Fig. 5b).
a, Schematic of rLAMP multiplexing with SARS-CoV-2 (N gene) and human internal control (RNase P) primer sets. b, DISCoVER signal of SARS-CoV-2-positive saliva samples after multiplexed rLAMP. Values are mean ± s.d. with n = 3 technical replicates. c, Fold change in DISCoVER signal relative to NTC on 30 negative clinical nasal samples at 5 min of Cas13 detection. d, Fold change in DISCoVER signal relative to NTC on 33 positive clinical nasal samples at 5 min of Cas13 detection.
Next, we validated DISCoVER in total RNA extracted from patient respiratory swab samples. Leftover RNA from 30 negative specimens and 33 positive specimens (Ct values 13–35) that had previously been tested by RT-qPCR in a CLIA testing lab37 were used as input for the DISCoVER benchtop assay. (Fig. 5c,d). We were able to correctly call 30 out of 30 negative samples within 5 min of Cas13 detection: no N gene was detected in these samples, while RNase P signal was robustly detected. Importantly, we detected the N gene in 31 out of 33 positive samples within the same timeframe. For the two false-negative specimens, RNaseP was detected but N gene was not, suggesting a difference in LOD between the two assays. On the basis of the clinical sample dataset, we report the sensitivity of DISCoVER on extracted RNA to be 93.9% with 100% specificity. Additionally, we observe the PPV to be 100%, with an NPV of 93.75%.
Finally, we developed a compact and portable microfluidic device to perform the assay in a setting leading to point-of-care applications. The chemistry was adapted to run on a gravity-driven microfluidic cartridge, with temperature-controlled reaction and detection chambers (Fig. 6a). The device contains air displacement pumps for controlled fluidic flow, a resistive heater for the rLAMP chamber, and a thermoelectric cooler/heater (TEC) for Cas13 chamber cooling and heating (Fig. 6b). The assay fluorescence detection was performed by a compact custom detector for real-time sample readout.
a, Left: illustration of the final instrument in which the cartridge is inserted. Inset shows picture of the entire microfluidic gravity-driven cartridge. Right: images of the sample metering and rLAMP reaction chamber (1), rLAMP metering and Cas13 mixing chamber (2) and the detection chamber (3). b, Front (left) and rear (right) illustration of the instrument components, including detection system, heaters, air-driven valves and mechanical valves. c, Schematic of the cartridge function. The reaction can be separated in four steps: saliva metering (1), amplification reaction (2), post-amplification metering + Cas13 mix (3), and Cas13 reaction in detection chambers (4). Reagents stored on the cartridge are separated via a proprietary hydrophobic solution to avoid premature initiation of the reactions. After the LAMP reaction, the sample can be split into two reactions: the left part of the cartridge will expose the sample to N gene crRNA (step 3a), while the right side of the cartridge will act as internal control with only RNase P crRNA (step 3b). Saliva samples went through only step 3a, while clinical samples went through steps 3a and 3b.
A microfluidic cartridge was developed for positive or negative saliva sample detection (Fig. 6c). In the first iteration of cartridge assay design, inactivated (75 °C for 30 min) positive commercial saliva sample is loaded onto the cartridge and actively metered into the rLAMP chamber, where it is mixed with N gene primers and rLAMP master mix. The solution is then heated to 65 °C for 30 min while the Cas13 reagents are maintained at 25 °C with the TEC. At the end of the rLAMP reaction, the active valve below the chamber opens, enabling gravity-assisted flow of 4 μl of the amplified solution into a metering channel. Only the left channel (Fig. 6c, step 3a) was open, allowing for Cas13 reagents and its N gene-targeting CRISPR RNA (crRNA) to be pushed through the metering channel into the mixing chamber, where it was then heated to 37 °C for 10 min. The right channel (Fig. 6c, step 3b) was added in the later version of the cartridge. Cas13 mixed with the rLAMP solution was aspirated to the detection chamber, and the reaction was monitored using a fluorescence detector. Microfluidic actuation times were ~8 min across the entire on-board assay. We demonstrate the detection of a dilution series from 200 to 40 copies μl−1 of SARS-CoV-2 genomic RNA in whole saliva, with a start-to-end (including inactivation step) reaction time of 78 min (Fig. 7a). This indicates the potential point-of-care application of this microfluidic system, where DISCoVER can detect SARS-CoV-2 RNA from saliva samples with attomolar sensitivity.
a, Heat map depicting the fold change in DISCoVER signal on negative and positive saliva samples relative to the NTC. b, Heat map depicting the fold change in SARS-CoV-2 RNA positive and negative clinical samples from nasal swabs, relative to NTC. Ct values for N gene target by qPCR detection is listed on the heat map axis. c, Graph depicting raw fluorescence over time for both detection chambers (blue for RNase P and red for N gene) in NTC (left), negative (middle) and positive (right, Ct 16) samples. Inset within each plot is fluorescent images of detection chamber for NTC cartridge (no viral sample), negative and positive clinical sample (with RT–qPCR Ct ranging from 16 to 21). The left detection chamber is specific for N gene detection, whereas the right chamber is specific for RNase P. d, Results of on-board DISCoVER on ten clinical saliva samples and three negative clinical saliva samples. Fold increase is calculated over a blank cartridge. A two-fold fluorescence increase over blank cartridges at 50 min was the experimentally determined criterion for positive results. Samples with value below this threshold at 5 min were declared negative.
Next, we optimized the cartridge design by multiplexing the detection of both N gene and RNase P during the rLAMP step to add an internal process control (step 3b in Fig. 6c). Multiplexed rLAMP was performed before the amplified reaction was split into two Cas13 detection chambers, each containing a single crRNA targeting either the N gene (left chamber, step 3a) or RNase P (right chamber, step 3b) rLAMP amplicon (Fig. 6c).
We then re-tested RNAs from three negative and three positive (Ct 16–21) respiratory clinical samples on the full DISCoVER device (Fig. 7b). We correctly detected SARS-CoV-2 in all three positive specimens (Fig. 7c, bottom), and detected only RNase P signal for all three negative specimens (Fig. 7c, middle). NTC cartridges without any viral sample were run initially to determine the baseline fluorescence, enabling us to determine the fold change of a positive sample from baseline (Fig. 7c, top).
To demonstrate the robustness and reproducibility of this system, we ran an additional 25 contrived samples on the DISCoVER device (20 contrived positive samples at 500 copies μl–1 and 5 expected negative samples at 0 copies μl–1) (Extended Data Fig. 2). To enable a robust process control across contrived specimens made from commercial saliva samples, we optimized multiplexed amplification off-board and on-board by using human ACTB (β-actin) gene primers in the rLAMP step instead of RNase P primer (Extended Data Fig. 2a,b). For the contrived saliva samples, we were able to shorten the inactivation step to 5 min at 95 °C and designed a simplified cartridge without the metering step (Extended Data Fig. 2c) for shortening the overall assay time to about 50 min, including sample inactivation38 and microfluidic actuation time. Samples with on-board signal at least two-fold change over background were called positive, and samples that did not reach that threshold were considered negative (Extended Data Fig. 2d). We then validated the device on ten natural, field-collected clinical saliva samples from individuals who tested positive by RT–qPCR for COVID-19 and three natural clinical saliva samples from individuals who tested negative (Fig. 7d and Supplementary Table 5). Owing to Environmental Health and Safety guidelines regarding field-collected saliva samples, the sample inactivation time was extended to 20 min at 95 °C, bringing the total assay time to 63 min for this validation. All 13 field-collected clinical saliva samples and 25 contrived commercial samples were correctly detected with attomolar sensitivity on a fully automated microfluidic system with minimal user interaction (Supplementary Video).
Discussion
We developed an RNA-extraction-free detection workflow, demonstrating the attomolar detection of SARS-CoV-2 RNA in unextracted saliva. The DISCoVER assay combines a 30-min rLAMP step followed by T7 transcription and Cas13-based detection, creating a rapid testing protocol with attomolar sensitivity and high specificity. As each modified LAMP product can serve as a substrate for transcription initiation, maximum Cas13 signal is reached in under 5 min owing to rapid generation of nanomolar substrate concentrations (Supplementary Fig. 3). The combination of sensitive nucleic acid amplification with CRISPR-mediated specificity and programmability has the potential to enable flexible diagnostics for diverse pathogen detection.
We demonstrated the DISCoVER system first on unextracted saliva, with the aim of enabling the key steps for a point-of-care diagnostic. As a saliva-based assay may not require medical personnel for sample collection, it is preferable to NP swabs, and the increased comfort of sample collection will probably incentivize patient compliance and commitment to frequent testing. It has also been shown that saliva is a reliable sample matrix for asymptomatic testing in community surveillance, as saliva samples have comparable viral titres to NP swabs11. To diversify the sampling supply chain, DISCoVER can also be applied to other samples such as NP swabs and self-administered nasal swabs.
In comparison with other CRISPR detection methods such as DETECTR and STOPCovid V2, DISCoVER does not employ a sample extraction or purification step39,40, eliminating reliance on commercial RNA extraction kits while maintaining comparable sensitivity. The direct lysis method employed here exploits common reagents for chemical reduction and ion chelation that are simple, widely available at low cost and stable at room temperature. DISCoVER maintains attomolar sensitivity compared with other CRISPR-based detection assays that include viral RNA extraction and purification20,39,41.
We further validate the ability of DISCoVER to detect multiple variants of SARS-CoV-2, specifically the B.1.1.7 variant of concern, which has been of broad medical interest owing to its potential increased transmissibility36. With additional development of mutant-specific DISCoVER probe sets, it may be possible to detect one variant of concern over another more selectively. Here we optimized for pan-SARS-CoV-2 surveillance, using primers designed to detect as many SARS-CoV-2 variants as possible. To further test DISCoVER specificity, we assayed for cross-recognition of viral genomic RNA from other common respiratory pathogens, including influenza A H1N1 and H3N2, influenza B and human coronavirus OC43. We did not observe a discernible effect on DISCoVER kinetics or specificity.
We also demonstrate multiplexed SARS-CoV-2 detection with a human internal control during the DISCoVER amplification stage. This could be extended for multi-colour detection using additional Cas enzymes with orthogonal cleavage motifs, each acting on specific reporters for distinct fluorescent channels42. Higher-order multiplexing can be further developed for influenza types A and B and other common respiratory viruses that would be desirable to detect in a single test. The inherent ability of the core enzymes in DISCoVER to convert between any RNA and DNA sequence implies that any pathogen that is inactivated and lysed by our protocol can be detected.
We validated DISCoVER on 33 positive and 30 negative patient samples, reporting a PPV of 100% and an NPV of 93.75% for total RNA extracted from nasal swabs. We observe 100% specificity and 93.9% sensitivity compared with qPCR testing for samples with Ct values ranging from 13 to 35. Finally, we demonstrate the integration of DISCoVER with a microfluidic system and detection device. With simple heating and cooling elements and air displacement pumps, the gravity-driven microfluidic cartridge can perform the DISCoVER workflow on both contrived positive saliva and patient samples in <50 min, including inactivation, with a real-time fluorescence readout. Further development and deployment of this assembly shows potential towards a point-of-care system, which will greatly facilitate frequent, on-site molecular diagnostics.
Methods
CRISPR–Cas direct nucleic acid detection
For Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a), crRNA and activator DNA were synthesized via Integrated DNA Technologies (IDT). Detection with Cas12a was performed using 1× NEB 2.1 buffer (B7202S), 100 nM LbCas12a protein, 100 nM crRNA, varying concentrations (100 nM, 100 nM, 10 nM, 1 nM, 100 pM and 10 pM) of double-stranded DNA activator and 200 nM DNaseAlert (IDT). LbCas12a and crRNA was pre-mixed at 10× concentration at a 1:1 molar ratio and incubated at room temperature for 15 min before adding it to the reaction. The reaction was heated at 37 °C with 15-s interval detection with a Tecan Spark multimode microplate reader. For background subtraction, the fluorescence values of the no-activator reaction were subtracted from the fluorescence values of sample reactions at each timepoint. Maximum fluorescence was obtained by mixing DNase I and 200 nM DNaseAlert with 1× NEB 2.1 buffer. This saturated maximum fluorescence value was divided in half; time to reach this half-maximum fluorescence was calculated for each concentration of activator.
For Leptotrichia buccalis Cas13a (LbuCas13a), the crRNA and the activator were commercially synthesized from Synthego. Detection with Cas13a was performed using 1× Cas13 reaction buffer, 100 nM LbuCas13a protein, 100 nM crRNA, varying concentrations (100 nM, 100 nM, 10 nM, 1 nM, 100 pM and 10 pM) of single-stranded RNA and 200 nM Cas13 reporter (IDT). LbuCas13a and crRNA was pre-mixed at 10× concentration in 1:1 ratio and incubated at room temperature for 15 min before adding it to the reaction. The rest of the reaction, and time to half-maximum fluorescence, was performed or analysed as described above. Maximum fluorescence was obtained by mixing RNase A and 200 nM Cas13 reporter with 1× buffer. 5× Cas13 reaction buffer (pH 6.8) is composed of 100 mM HEPES–Na pH 6.8 (Sigma), 250 mM KCl (Sigma), 25 mM MgCl2 (Sigma) and 25% glycerol (Thermo Fisher).
LbuCas13 protein production
The expression and purification of Leptotrichia buccalis Cas13a (LbuCas13a) was performed as previously described24 with modifications, summarized here: His6–MBP–TEV-tagged Cas13a was prepared in Escherichia coli Rosetta2 (DE3) grown in TB at 37 °C. At an OD600 of 0.6–0.8, cultures were cooled on ice for 15 min before induction with 0.5 mM isopropyl β-d-1-thiogalactopyranoside and expression overnight at 16 °C. Cell pellets measuring close to 10 ml in a 50 ml Falcon tube were re-suspended in 100 ml of lysis buffer each. Soluble His6–MBP–TEV Cas13a was clarified by centrifugation at 15,000g, then loaded onto a 5 ml HiTrap NiNTA column (GE Healthcare) and eluted over a linear imidazole (0.01–0.3 M) gradient via fast protein liquid chromatography (ÄKTA Pure). Following overnight dialysis and TEV digestion, LbuCas13a underwent purification by ion exchange and removal of MBP (5 ml HiTrap SP, MBP trap HP, GE Healthcare). Finally, size-exclusion chromatography was performed (S200 16/60, GE Healthcare) with 1 mM TCEP supplemented into the gel filtration buffer, and peak fractions were pooled, concentrated and aliquoted into PCR strip tubes, then snap frozen in liquid nitrogen.
LbCas12a protein production
The expression and purification of Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a) was performed as previously described with the addition of 1 mM EDTA in the dialysis buffer to prevent protein precipitation during His-tag removal by TEV protease43.
LAMP and rLAMP reactions
LAMP and rLAMP DNA amplification reactions were conducted with 2× LAMP Mastermix (NEB, E1700), 0.4 µl LAMP dye (NEB, E1700), 2 µl of 10× primer mix (IDT; 16 μM of FIP/BIP, 4 μM FLoop/BLoop and 2 μM of F3/B3), and 7.6 µl of sample. Eight microlitres of sample was added to reactions in which no dye was added. For multiplexed LAMP, 1 μl of RNase P primer with T7 promoter on the BIP 5′ primer was added to the 20 μl reaction. Detection was performed at 65 °C for 30–60 min in a CFX96 Touch Real-Time PCR machine (Bio-Rad) or a QuantStudio 3 Real-Time PCR System (Thermo). Time to threshold was calculated using single threshold analysis mode. LOD for LAMP was determined as the concentration where there was amplification observed with at least 95% of the samples. LAMP amplification reactions for clinical samples were conducted with 2× LAMP Mastermix, 2 µl of 10× N gene primer mix, 1 µl of RNase P primer mix and 5 µl of sample in a 20 µl reaction.
Twist Synthetic SARS-CoV-2 RNA Control 2 (Twist Bioscience, SKU 102024) was used as a template to perform the primer screen and establish LOD ranges. The volume of template to be used and the final concentration of template in the reaction was calculated on the basis of the initial concentration provided by the vendor as 106 copies µl−1.
rLAMP assay development
Restriction digestion of rLAMP DNA products was performed on 3 µl of amplified rLAMP product using 2 µl of AfeI (NEB R0652L), 2 µl 10× CutSmart Buffer and 13 µl of water. This 20 µl of reaction was heated at 37 °C for 30 min. Undigested and digested rLAMP products were treated with 4 µl 1 U µl−1 RQ DNase I and 1 µl of 10× reaction buffer (Promega, M6101). The reaction was heated at 37 °C for 30 min, and 1 µl of STOP solution (Promega M6101) was added, followed by incubation at 65 °C for 10 min. rLAMP products were analysed using 15% TBE–urea gels (Thermo Fisher, EC68855BOX).
Direct saliva lysis
SARS-CoV-2-negative saliva (Lee Biosolutions, 991-05-S-25) was used to check compatibility of saliva with the DISCoVER workflow. All saliva samples were collected before November 2019 as per vendor. When viral synthetic RNA was used, Twist Synthetic SARS-CoV-2 RNA Control 2 (SARS-CoV-2 isolate Wuhan-Hu-1) was spiked in the commercial saliva and used as a template for mock samples. For LOD determination, SARS-CoV-2 viral seedstocks were spiked in negative saliva background under BSL-3 containment and used as a template for mock samples.
Commercial saliva was mixed with lysis reagents and heated at 75 °C for 30 min or 95 °C for 5 min using a heat block. Twist Synthetic RNA was added after completion of heat inactivation of saliva. Viral seedstocks were spiked in saliva in the BSL3 facility before heat inactivation. Inactivating reagent 1 was 2.5 mM TCEP/1 mM EDTA at 1× concentration when mixed with saliva. Inactivating reagent 2 was 100 mM TCEP/1 mM EDTA at 1× concentration when mixed with saliva. Inactivating reagent 3 was Quickextract DNA (Lucigen QE09050), inactivating reagent 4 was Quickextract RNA (Lucigen QER090150) and inactivating reagent 5 was RNA/DNAShield (Zymo 76020-420).
Field-collected, clinical saliva samples from the Salinas Valley farmworkers study were lysed in a bead bath set to 95 °C for 20 min. According to ref. 37, the inactivation time used for contrived samples made from commercial saliva can be used here as well, but we were unable to test this owing to restrictions placed by UC Berkeley Environment, Health, and Safety (EH&S) on clinical sample inactivation protocols.
LOD determination
To determine the LOD, SARS-CoV-2 (USA-WA1/2020; BEI Resources, NR-52281) viral stocks amplified once in Vero-E6 cells (ATCC) was diluted in medium (DMEM + 10% FBS + 1% penicillin–streptomycin) to obtain various concentrations, spiked into saliva and inactivated by adding 1× of low TCEP/EDTA inactivating reagent and heating the samples at 75 °C. The dilutions of virus in medium were inactivated by heating at 75 °C for 30 min to comply with EH&S requirements (UC Berkeley) and RT–qPCR was performed using Luna Universal Probe One-Step RT-qPCR Kit (NEB E3006) and E gene-targeting primers to determine the final concentration of viral RNA. Synthetic genomic RNA fragments (Twist Synthetic SARS-CoV-2 RNA Control 2) were used to obtain a standard curve for the calculations. The mock sample was detected as positive if the fold change (ratio of fluorescence value of sample to fluorescence value of no template control) is greater than 2 within 5 min. LOD for the mock samples was determined as the concentration where there was detection with at least 95% of the samples.
Clinical sample RNA extraction
RNA extracted from respiratory swab samples was obtained from the Innovative Genomics Institute (IGI) Clinical Laboratory. These specimens were collected for SARS-CoV-2 diagnostic purposes, and the LuNER assay37 had been previously performed on them. In brief, respiratory specimens were collected from patients (from the oropharynx, anterior nares or mid-turbinate) by a healthcare provider. RNA was extracted in the IGI Clinical Laboratory using an automated workflow based on the MagMAX Viral/Pathogen Nucleic Acid Isolation Kit (Thermo Fisher A42352) and then assayed for N and E genes and RNaseP. Specimens were de-identified and extracted RNA was provided with clinical Ct values for the experiments described here.
Point-of-care validation with saliva samples
We determined that a two-fold change between a positive signal and negative control at 5 min into Cas13 detection is sufficient to distinguish contrived positive and expected negative saliva samples. This experimentally derived threshold was used for calling positives and negatives in clinical samples. A sample is determined to be positive if both N gene and process control signal fold change is above two-fold within 5 min. A sample is determined positive if N gene shows two-fold change within 5 min, even if the process control does not show two-fold change, as this could indicate loss or degradation of human cellular material in saliva. A sample is determined to be negative if the process control shows a >2-fold change within 5 min while N gene signal does not appear. If both N gene and the process control fold change are below 2, the test is considered invalid.
Cas13a guide design pipeline
A LbuCas13a guide targeting SARS-CoV-2 was designed by prioritizing sensitivity, specificity and predicted secondary structure. Twenty-six candidate spacers of 20 nucleotides were identified within the N set 1 amplicon region of the SARS-CoV-2 reference genome (wuhCor1), targeting both the forward and reverse strands. Sensitivity was determined by pairwise aligning available SARS-CoV-2 genomes from GISAID with the reference genome and calculating the number of mismatches between the sample and reference genome for each candidate spacer. The percentage of SARS-CoV-2 genomes detected by each spacer, with no more than one mismatch, was then calculated, and spacers matching less than 99% of SARS-CoV-2 genomes were discarded. To ensure specificity, candidate spacers that were determined to be sensitive were aligned to other reference genomes of human coronaviruses, specifically, SARS, MERS, 229E, NL63, OC43 and HKU1. Spacers that matched any of these other coronaviruses with two or fewer mismatches were discarded. The remaining spacers were also aligned to the human transcriptome, again allowing for two mismatches, and spacers with any alignments were discarded to ensure the guide would not be complementary to off-target human transcripts. To ensure that the spacer sequences would allow for Cas13a binding to the direct repeat scaffold, the concatenated repeat and spacer sequence was folded using RNAfold and evaluated for correct hairpin structure in the direct repeat and single strandedness in the spacer sequence. Out of five possible SARS-CoV-2 spacers passing our sensitivity, specificity and structure criteria, one was chosen for use as a guide RNA. A control guide targeting RNase P was selected with the same criteria to avoid matching SARS-CoV-2 and human transcripts and to ensure proper RNA structure. In the validation experiment (Fig. 7d and Extended Data Fig. 2d), owing to the difference in saliva composition between commercial and clinical samples, the multiplexed amplification was optimized off-board and on-board by using human ACTB gene primers into the rLAMP step instead of RNase P primer. ACTB guides were chosen using the same pipeline as N set 1 and RNase P.
T7 transcription
Transcription was performed on amplified rLAMP product using 2 µl of 1 mg ml−1 of T7 polymerase, 4 µl of 100 mM NTP mix (NEB N0450), 1 µl of 200 mM DTT, 4 µl of 5× transcription buffer, 2 µl of LAMP product and 7 µl of water. This 20 µl of reaction was heated at 37 °C for 30 min. The 5× transcription buffer is composed of 150 mM Tris–Cl, pH 8.1 at room temperature, 125 mM MgCl2, 0.05% Triton X-100 (Sigma Aldrich, X100) and 10 mM spermidine and stored at −20 °C.
Cas13 detection
Cas13 detection was performed as a 20 µl reaction using 2 µl of 1:100 diluted transcription product of LAMP, 1 µl 5× Cas13 reaction buffer, 2 µl of 2 µM Cas13 reporter, 2 µl of 10× ribonucleoprotein (RNP), 13 µl of 1× Cas13 reaction buffer. The 10× RNP was made as a 2:1 ratio of Cas13:crRNA with final concentration of Cas13 as 20 nM and incubated at room temperature for 15 min before adding it to the reaction. This 20 µl reaction was heated at 37 °C and read every 30 s in TECAN Spark multimode microplate reader using FAM channel (excitation 485 nm, emission 535 nm).
One-pot T7 transcription and Cas13 detection
One-pot T7 and Cas13 was performed by combining 2 µl of 1 mg ml−1 of T7 polymerase, 20 mM NTP mix, 10 mM DTT and 25 mM MgCl2 with the Cas13 reaction mix. Two microlitres of 1:100 dilution of rLAMP product is used for 20 µl of one-pot T7–Cas13 reaction. For testing clinical samples, 2 µl of 1× rLAMP product is used for 20 µl of one-pot T7–Cas13 reaction.
DISCoVER specificity testing
Synthetic viral RNA of Influenza A H1N1 (Twist Bioscience, SKU 103001), influenza A H3N2 (Twist Bioscience, SKU 103002), influenza B (Twist Bioscience, SKU 103003) and human coronavirus OC32 (Twist Bioscience, SKU 103013) was spiked into inactivated saliva (Lee Biosolutions, 991-05-S-25), with and without Twist Synthetic SARS-CoV-2 RNA Control 2. rLAMP and one-pot T7 transcription as described above were performed with the N gene crRNA.
Synthetic viral RNA of SARS-CoV-2 variant B.1.1.7_710528 Control 14 (Twist Bioscience, SKU 103907) and SARS-CoV-2 variant B.1.1.7_601443 Control 15 (Twist Bioscience, SKU 103909) was spiked into inactivated saliva (Lee Biosolutions, 991-05-S-25). rLAMP and one-pot T7 transcription as described above were performed with the N gene crRNA.
Instrumentation of point-of-care system
A prototype instrument was designed and fabricated by Wainamics Inc. The instrument was designed specifically to demonstrate feasibility of the system and with features that would allow for an easy transition to a manufacturable end-use instrument. A combination of off-the-shelf components, custom-designed components (designed in SolidWorks), a custom electronic board based around a Teensy 3.2 microcontroller, and custom software (written in C) were integrated together to fabricate a prototype instrument. The system was intended to demonstrate feasibility of the assay on-board a prototype thermoplastic device in an automated manner. Care was taken to ensure that all design elements would be transferable to commonly used high-volume manufacturing methods (for example, die-cutting, injection moulding, etc).
Custom-designed components for the prototype system were fabricated using common manufacturing methods including commercial computer numerical control (CNC) machining (Protolabs) of aluminium and Delrin components, fused deposition modeling (FDM) 3D printing (Dremel 3D45) and stereolithography (SLA) 3D printing (Formlabs Form3). The commercially available components used consisted of a Norgren Kloehn V6c syringe pump with eight-way rotary valve, Burkert Type 6724 three-way solenoid valves, a Laird Thermal Systems temperature controller (TC-XX-PR-59) controlling a Marlow Industries TEC unit (NL2064T-11AB), control fans (Sunon Fans MF25101V1-1000U-A99) and resistive cartridge heaters (E3D, 12 V 40 W). A custom graphical user interface on a Windows 10 PC allowed the user to control various functionality of the system and cartridge (that is, actuate values, control volumes of air displaced into cartridge by pump, control flow rate of displaced air, control heater temperatures, etc). Script-recording allowed the user to generate and record instrument system scripts that were used to automatically run the assay on-board the cartridge to ensure precision and consistency across cartridge assay runs.
Fluidic actuation time was measured to be ~8 min. A key element of the instrument is a custom manifold that forms an airtight seal to the plastic cartridge when engaged, allowing actuation of liquid inside the cartridge. The manifold consists of seven ports connected with commercial valves to a single custom syringe pump. The ports are sealed to the cartridge using gaskets that are built into the manifold. On the plastic cartridge, each of the ports is isolated from the instrument by a hydrophobic membrane that allows passage of air but not aqueous fluids, therefore protecting the instrument from contamination by any assay reagent or saliva. When positive air pressure or negative pressure is applied to the appropriate manifold ports, the pressure will push or pull liquid within the cartridge according to flow rate and volume of air displaced. The valves on the manifold can also vent to the atmosphere, allowing pressure inside the cartridge to equilibrate. The volume of air displaced and pump flow rates are adjusted to allow for thorough mixing of reagents and minimum residual volume loss inside channels.
The overall prototype instrument measures approximately 12′ wide, 18′ deep and 12′ tall. A custom-designed instrument with custom electronic board, custom manifold with built-in pumps and valves and custom optics could be as small as 8′ × 9′ × 9′.
The resistive heater and TEC are adjusted by measuring temperatures reached inside the cartridge during operation using a cartridge with embedded thermocouples. The set temperature is usually 5–8 °C higher than the actual temperature inside the cartridge owing to heat transfer losses through the plastic. The setpoints for the heaters are set accordingly with a matching offset. For the cartridge used in the clinical testing, the LAMP reaction chamber heater was set to 72 °C and Cas13 detection chamber set to 39 °C.
Cartridge fabrication
The cartridge was designed by Wainamics Inc. in SolidWorks. Prototype cartridges were designed and fabricated with manufacturability in mind. Laser-cut polymethyl methacrylate layers of various thickness (0.25–1 mm, Clarex Precision from Nitto Jushi Kogyo Co.) were laminated together using PCR-compatible pressure-sensitive adhesives (Adhesive Research AR90880 and AR92712) to achieve the desired channel geometries. All channel geometries were designed to be easily translated to, and replicated by, mass-manufacturing techniques such as injection moulding and rotary die-cutting. Each layer was cut by CO2 laser (BossLaser), and the entire cartridge was hand-assembled by Wainamics Inc. Hydrophobic membranes (Sterlitech PTFE029025) were used to allow passage of gas, not liquid, to prevent contamination of the prototype instrument. Silicone rubber sheet (McMaster-Carr 1460N11) was used to fabricate actuatable cartridge valves to control solution flow.
Cartridge thermal calibration
A cartridge containing thermocouples (Omega Engineering) embedded in thermal epoxy (McMaster-Carr, #7548A11) was used for temperature calibration purposes. Power settings for resistive heaters and TEC were manually adjusted using the readings from the temperature calibration cartridge. A temperature calibration run using feedback from a thermal test cartridge is shown in Supplementary Fig. 5. The difference between the set temperature and the recorded temperature increases at higher temperatures (not stable). It is therefore important to calibrate the system by verifying the set temperature necessary to match the required reaction temperature.
Cartridge fluidics calibration
Owing to losses of reagents along the channels, multiple calibration experiments were performed to ensure correct reaction volumes (Supplementary Table 3). To measure the amount of volume lost between the ‘starting reservoir’ and the ‘destination chamber’, the experimental protocol was the following: A given volume of fluorescent dye was loaded in the starting reservoir. On the other hand, a known volume of PBS with 0.1% Tween-20 was added to the ‘destination’ chamber. The experimental fluidic sequence was run to flow the dye from the reservoir to the chamber. Once mixed, the diluted dye in the chamber was withdrawn and its fluorescence was assessed via plate reader. On the plate reader, a known titration of the dye was simultaneously assessed to create the calibration curve. By comparing the cartridge-withdrawn dye fluorescence to the calibration curve, we could deduce the volume of the dye recovered after the fluidic flow and the amount of dye lost in the channels. A solution of 100 nM fluorescein (Sigma F6377) in 1× PBS (Sigma P5493) with 0.1% Tween-20 (Sigma P1379) was used for visual fluidic test purposes. FamT10 (IDT) in 1× PBS in various concentrations (0 nM to 200 nm) was also used as a calibrator solution for optical characterization of the cartridge with the detection system.
Detection device
For imaging the 40 μl sample volumes in the reaction chambers, we require a fairly large field of view (FOV) and a modest numerical aperture (NA) enabling sufficient depth of focus without images of adjacent sample wells overlapping. To accomplish this in a relatively low-cost, compact device, we designed a custom system using a pair of eyepieces (Edmund Optics, PN#66-208 and 66-210), yielding a system with NA 0.09, FOV diameter 12.0 mm, and magnification of 0.54 (chosen to match the sensor size of the Thorlabs CS165MU1 camera to the FOV; sampling at ≥Nyquist is unnecessary in this ‘light-bucket’ application) (Supplementary Table 4). The overall system is compact, with nominal track length (sample to camera) of ~75 mm. Fluorescence filters were from Chroma Technologies, ET470/40x, T495lpxr and ET535/70m, with excitation provided by a 965 mW, 470 nm LED (Thorlabs M470L4 driven by an LEDD1B), providing a maximum of ~225 mW into the 12 mm diameter sample FOV in an epi-illumination Kohler geometry. Custom control of the imaging hardware was implemented in MATLAB (2020a), using Thorlabs drivers and SDK (ThorCam) to control the camera acquisition, and serial communication to an Arduino Bluefruit Feather board to electronically trigger the LED illumination in synchrony with camera image acquisition.
The multiplex cartridge has two detection chambers for both N gene and internal control (RNaseP or ACTB). Every day, before running the assay on saliva samples, a blank cartridge filled with buffer as well as a fluorescent slide were imaged for background and illumination uniformity corrections, respectively. One region of interest (ROI) (150 × 400 pixels) was selected in each chamber, and the blank cartridge was imaged at the same gain and exposure time as subsequent experiments (2 dB and 150 ms). The intensity measured in each ROI was used to normalize the sample images to background. To correct for the illumination non-uniformity, a clean fluorescent slide was imaged at a gain of 1 and exposure of 0.1 s.
Both background and illumination uniformity correction were performed using a prospective correction method44. In short, the measured intensities for each pixel, x, of reaction chambers is related to actual intensities through
where S is a linear scaling factor that models distortions to an image due to non-uniform illumination; B is illumination-dependent background signal that accounts for scattering and background fluorescence from the sample chambers; and D is an additive zero-light term, which is due to camera offset and fixed pattern thermal noise. S, B and D were determined from images of a blank cartridge and a uniform fluorescent slide acquired before experiments at the start of each day. Experimental images were processed according to equation (1) to retrieve corrected sample signals. A custom MATLAB script was developed to control the illumination and detection parameters exposure time, gain, time interval between each image acquisition and total number of frames to capture. The script also allowed live preview of the cartridge for optimal alignment and selection of ROIs inside the reaction chambers. Once the assay was started, the script also automatically triggered the image acquisition as the reaction mix was pumped into the detection chambers. At this point, the script displayed images captured at each timepoint and the corresponding mean intensity of the selected ROIs during the experiment.
Commercial saliva assay on microfluidic cartridge
A proof-of-concept system was initially developed with a single reaction channel for positive or negative N gene detection. Commercial saliva (Lee Biosolutions, 991-05-S-25), spiked with Twist Synthetic SARS-CoV-2 RNA as positive sample or with water for negative sample, was used on this system for instrument calibration and rapid outcomes. As shown in Fig. 5, the reaction can be separated into four steps: (1) saliva metering, (2) LAMP reaction, (3) LAMP metering + Cas13/N gene crRNA mix and (4) Cas13 reaction in detection chamber. Owing to losses of reagents along the channels, multiple calibration experiments were performed as described earlier, to ensure correct reaction volumes (as referred here; loading volumes can be found in Supplementary Table 3). In step 1 (top part), 16 μl of saliva is actively aspirated by syringe pump into the metering channel. The syringe pump can then actively push the primer (4 μl) and master mix (20 μl), separated via a proprietary hydrophobic solution (35 μl), through the metering channel into the LAMP mixing chamber. An additional 50 μl of air (at 250 μl min−1) is pushed into the chamber to allow for air bubbles to mix the LAMP reaction (step 2). The resistive heater is turned on to heat the LAMP chamber to 65 °C for 30 min and allow for the reaction to happen. Simultaneously, the TEC is set at 25 °C to maintain Cas13, T7 mix, fluorescence quencher (FQ) and crRNA reagents at room temperature. After the LAMP reaction (step 3), the lower active valve opens, and gravity will drive 4 μl of the LAMP product to flow through the left metering channel. The syringe pump actively pushes 32 μl T7mix + FQ with 4 μl Cas13 + N gene crRNA through the metering channels into the left mixing chamber. An additional 50 μl of air is pushed into both chambers for mixing. In step 4, 45 μl of the solution is actively aspirated into the respective detection chambers. The TEC is turned up to 37 °C, and fluorescence images are acquired every 15 s for 15–30 min.
Clinical sample assay on multiplex system
Once the cartridge detected positive and negative saliva samples, we designed a new cartridge for clinical extracted RNA detection. These samples were leftover clinical extracted specimen from the Clinical Laboratory Improvement Amendments (CLIA) testing lab. As we could access only 5 μl per clinical sample, the saliva metering fluidics (step 1 in Fig. 5c) were not used. Instead, the clinical sample was directly loaded in the LAMP chamber. The reaction can be separated in three following steps of Fig. 5c: (2) LAMP reaction, (3) LAMP metering + Cas13/crRNA mix and (4) Cas13 reaction in detection chambers. We also multiplexed step 3 and step 4 so that the left part of the cartridge will expose the sample to N gene crRNA (step 3a) while the right side of the cartridge will act as internal control with RNase P crRNA (step 3b), to show that enough cellular material was present in the sample. Below we report reaction volumes; loading volumes can be found in Supplementary Table 3. In step 2, the 5 μl of clinical sample is directly loaded into the LAMP chamber. The syringe pump can then actively push the primer (4 μl) and master mix (20 μl), separated via a proprietary hydrophobic solution (35 μl), into the LAMP mixing chamber. An additional 50 μl of air is pushed into the chamber to allow for air bubbles to mix the LAMP reaction. The resistive heater is turned on to heat the LAMP chamber to 65 °C for 30 min and allow for the reaction to happen. Simultaneously, the TEC is set at 25 °C to maintain Cas13 and crRNA reagents at room temperature. Step 3 will split the LAMP sample into two reactions: the left part of the cartridge will expose the sample to N gene crRNA (step 3a), while the right side of the cartridge will act as internal control with only RNase P crRNA (step 3b). After the LAMP reaction, the two lower active valves open and gravity will drive 4 μl of the LAMP product to flow through left and right passive metering channels. The syringe pump actively pushes 32 μl T7 mix + FQ with 4 μl Cas13 + crRNA (N gene in left reservoir and RNase P in right reservoir) through the metering channels into the respective N gene (left) and RNase P (right) chamber. An additional 50 μl of air is pushed into both chambers for mixing. At this point, 45 μl of the solution is actively aspirated into the respective detection chambers (step 4). The left chamber will have LAMP product mixed with Cas13 and N gene, while the right chamber, internal control, will mix LAMP with Cas13 and RNase P crRNA. The TEC is turned up to 37 °C, and fluorescence images are acquired every 15 s for 15–30 min.
Contrived saliva assay on multiplex system
Owing to the difficulty of accessing large numbers of clinical samples, we demonstrated the robustness of the system by running saliva from distinct individuals (Lee Biosciences) where positive samples were generated by spiking Twist Synthetic SARS-CoV-2 RNA into the saliva matrix. Because of the different saliva composition of the commercial samples, we screened alternative process controls (Extended Data Fig. 2a) to address variable RNase P signal. First, we systematically tested the insertion of T7 promoter sequences in different regions of LAMP primers for ACTB, on both commercial saliva and RNA extract from HEK 293FT cells (Extended Data Fig. 2b), and performed a small guide screen to integrate it into the DISCoVER pipeline (Extended Data Fig. 2c). Owing to lower N gene signals in initial experiments relative to ACTB, we optimized the multiplexed amplification to consist of a reaction with 2:1 ratio of N gene to ACTB primer and addition of extra Bst 2.0 polymerase. We then ran 25 individual saliva samples, that is, 20 samples as contrived positives containing SARS-CoV-2 genomic RNA and 5 negative samples (Extended Data Fig. 2d) to demonstrate the reproducibility and robustness of this workflow.
The design of the cartridge was simplified to allow for more compact design, improved mixing and volume precision (Extended Data Fig. 2d). The reaction can be separated into the following steps: (1) LAMP reaction, (2) LAMP metering + Cas13/crRNA mix and (3) Cas13 reaction in detection chambers. Step 2 and step 3 were multiplexed so that the left part of the cartridge will expose the sample to N gene crRNA (step 2a) while the right side of the cartridge will act as internal control with ACTB crRNA (step 2b), to show that enough cellular material was present in the sample. In step 1, 18 μl of saliva sample is directly loaded into the sample chamber. The syringe pump could actively push a proprietary hydrophobic solution (30 μl) as well as the primer (10 μl) and master mix (40 μl), separated via some more hydrophobic solution (15 μl), into the LAMP mixing chamber. An additional 50 μl of air is pushed into the chamber to allow for air bubbles to mix the LAMP reaction. The resistive heater is turned on to heat the LAMP chamber to 63 °C for 45 min and allow for the reaction to happen. Some air is pushed every 15 min to mix the solution and improve the reaction outcome. Simultaneously, the TEC is set at 25 °C to maintain Cas13 and crRNA reagents at room temperature. Step 2 will split the LAMP sample into two reactions: the left part of the cartridge will expose the sample to N gene crRNA (step 2a), while the right side of the cartridge will act as internal control with only ACTB crRNA (step 2b). After the LAMP reaction, the two lower active valves open and gravity will drive 4 μl of the LAMP product to flow through left and right passive metering channels. The syringe pump actively pushes 40 μl T7 mix + FQ, 15 μl of hydrophobic solution and 10 μl Cas13 + crRNA (N gene in left reservoir and ACTB in right reservoir) through the metering channels into the respective N gene (left) and ACTB (right) chamber. An additional 50 μl of air is pushed into both chambers for mixing. At this point, 45 μl of the solution is actively aspirated into the respective detection chambers (step 3). The left chamber will have LAMP product mixed with Cas13 and N gene crRNA, while the right chamber, internal control, will also have the LAMP product but mixed with Cas13 and ACTB crRNA. The TEC is turned up to 37 °C, and fluorescence images are acquired every 10 s for 30 min. Fold change was determined between 5 and 10 min to call the assay outcome (Fig. 5g and Extended Data Fig. 2d).
Clinical saliva study and validation on multiplex system
Field-collected clinical saliva samples were collected from adults aged 18 years or older who resided in California and were receiving a COVID test (antigen, PCR or transcription-mediated amplification test) at Clinica de Salud del Valle de Salinas or any other Salinas Valley testing site (UC Berkeley IRB protocol number 2021-04-14268). Participants whose samples are discussed in this body of work included both agricultural and non-agricultural workers. Pregnant women were also invited to participate. Screening and enrolment took place in-person at the clinics conducting COVID-19 tests, as well as by phone for patients who had recently tested positive at Clinica de Salud del Valle de Salinas. For all recruitment and data collection, research and clinical staff wore masks and gloves. For in-person collection, field researchers provided the participant with an open-mouth container and asked them to spit in it up to a pre-determined mark of ~3 ml. The participant was then given a cover for the container, a disinfectant wipe and a paper towel. The participant was then asked to tightly cap the container and wipe the full outside of the container with the wipe. The field researcher, wearing gloves and other personal protective equipment, received the container, labelled it with a sticker containing the participant’s identification number and the date of collection, and placed it in a cooler with ice packs. For home-based recruitment of COVID-positive individuals, the field researchers phoned patients who had tested positive and determined their interest in participating in a research study. The field researcher then delivered a sample collection kit to the participant’s home. The kit included visual and written instructions for sample collection as well as a pre-labelled open-mouth container. The field researcher guided the participant through the sample collection via phone. The field researcher retrieved the kit from the participant upon completion and placed the saliva sample in a cooler with ice packs. Samples were then stored at −80 °C until transport to UC Berkeley on dry ice, and subsequently kept at −80 °C again. All samples were collected under the ethical oversight of the UC Berkeley Committee for Protection of Human Subjects.
When ready to test, all 66 samples available to us from the study were thawed and heat-inactivated for 20 min at 95 °C under university EH&S guidelines in BSL-2, despite 5 min being sufficient for saliva inactivation37. As the saliva samples themselves had not previously been tested, RT–qPCR was performed on each sample to obtain a positivity profile of the sample set. RT–qPCR was performed using Luna Universal Probe One-Step RT-qPCR Kit (NEB E3006) on N-gene targeting primers to determine the final concentration of viral RNA, and ACTB targeting primers as a process control. From the 66 samples screened, 13 samples were chosen (10 positive, 3 negative) across a range of Ct values (Supplementary Table 5) to run on the DISCoVER microfluidic device to validate the device’s capacity to differentiate negative and positive samples that were already confirmed by a gold-standard assay in RT–qPCR. Each of these saliva donor samples was run in the microfluidic system according to the protocol described in the contrived samples workflow and analysed in the same manner with an observation of two-fold fluorescence increase over background cartridges as an indicator of positivity for each target (Fig. 5g).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All nucleic acid sequences used are provided in the manuscript or in the Supplementary Information. The raw data for the clinical samples are provided as source data. All raw and analysed datasets generated during this study are available for research purposes from the corresponding authors on reasonable request. Source data are provided with this paper.
Code availability
The code used to control the imaging hardware is available as Supplementary Information.
References
Lavezzo, E. et al. Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature 584, 425–429 (2020).
Wölfel, R. et al. Virological assessment of hospitalized patients with COVID-2019. Nature 581, 465–469 (2020).
Zhao, J. et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clinical Infectious Diseases 71, 2027–2034 (2020).
Vandenberg, O., Martiny, D., Rochas, O., van Belkum, A. & Kozlakidis, Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-020-00461-z (2020).
IGI Testing Consortium. Blueprint for a pop-up SARS-CoV-2 testing lab. Nat. Biotechnol. 38, 791–797 (2020).
Bloom, J.S. et al. Massively scaled-up testing for SARS-CoV-2 RNA via next-generation sequencing of pooled and barcoded nasal and saliva samples. Nat. Biomed. Eng. 5, 657–665 (2020).
Rabe, B. A. & Cepko, C. SARS-CoV-2 detection using isothermal amplification and a rapid, inexpensive protocol for sample inactivation and purification. Proc. Natl Acad. Sci. USA 117, 24450–24458 (2020).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR–Cas13. Science 360, 444–448 (2018).
Weickmann, J. L. & Glitz, D. G. Human ribonucleases. Quantitation of pancreatic-like enzymes in serum, urine, and organ preparations. J. Biol. Chem. 257, 8705–8710 (1982).
Iwasaki, S. et al. Comparison of SARS-CoV-2 detection in nasopharyngeal swab and saliva. J. Infect. 81, e145–e147 (2020).
Wyllie, A. L. et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med. 383, 1283–1286 (2020).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR–C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Gootenberg, J. S. et al. Nucleic acid detection with CRISPR–Cas13a/C2c2. Science 356, 438–442 (2017).
Chen, J. S. et al. CRISPR–Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360, 436–439 (2018).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Ramachandran, A. et al. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl Acad. Sci. USA 117, 29518–29525 (2020).
Ning, B. et al. A smartphone-read ultrasensitive and quantitative saliva test for COVID-19. Sci. Adv. 7, (2021).
Brogan, D. J. et al. Development of a Rapid and Sensitive CasRx-Based Diagnostic Assay for SARS-CoV-2. ACS Sensors 6, 3957–3966 (2021).
Li, S.-Y. et al. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 28, 491–493 (2018).
Fozouni, P. et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184, 323–333.e9 (2021).
Nagamine, K., Hase, T. & Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 16, 223–229 (2002).
Hardinge, P. & Murray, J. A. H. Reduced false positives and improved reporting of loop-mediated isothermal amplification using quenched fluorescent primers. Sci. Rep. 9, 7400 (2019).
Liu, T. Y. et al. Accelerated RNA detection using tandem CRISPR nucleases. Nat. Chem. Biol. 17, 982–988 (2021).
East-Seletsky, A., O’Connell, M. R., Burstein, D., Knott, G. J. & Doudna, J. A. RNA targeting by functionally orthogonal type VI-A CRISPR–Cas enzymes. Mol. Cell 66, 373–383.e3 (2017).
Park, G.-S. et al. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). J. Mol. Diagn. 22, 729–735 (2020).
Yu, L. et al. Rapid Detection of COVID-19 Coronavirus Using a Reverse Transcriptional Loop-Mediated Isothermal Amplification (RT-LAMP) Diagnostic Platform. Clinical Chemistry 66, 975–977 (2020).
Zhang, Y. et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. bioRxiv https://doi.org/10.1101/2020.02.26.20028373 (2020).
Lamb, L. E., Bartolone, S. N., Ward, E., & Chancellor, M. B. Rapid detection of novel coronavirus/Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) by reverse transcription-loop-mediated isothermal amplification. PLoS ONE 15, e0234682 (2020).
Notomi, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, 63e (2000).
Dao Thi, V. L. et al. A colorimetric RT-LAMP assay and LAMP-sequencing for detecting SARS-CoV-2 RNA in clinical samples. Sci. Transl. Med. 12, eabc7075 (2020).
Song, J. et al. Single- and Two-Stage, Closed-Tube, Point-of-Care, Molecular Detection of SARS-CoV-2. Anal. Chem. 93, 13063–13071 (2021).
Ostheim, P. et al. Overcoming challenges in human saliva gene expression measurements. Sci. Rep. 10, 11147 (2020).
Chin, A. W. H. et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe 1, e10 (2020).
Meldrum, O. W. et al. Mucin gel assembly is controlled by a collective action of non-mucin proteins, disulfide bridges, Ca2+-mediated links, and hydrogen bonding. Sci. Rep. 8, 5802 (2018).
Tabachnik, N. F., Blackburn, P. & Cerami, A. Biochemical and rheological characterization of sputum mucins from a patient with cystic fibrosis. J. Biol. Chem. 256, 7161–7165 (1981).
Davies, N. G. et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 372, 6538 (2021).
Stahl, E. C. et al. LuNER: multiplexed SARS-CoV-2 detection in clinical swab and wastewater samples. PLoS ONE 16, e0258263 (2021).
Batéjat, C., Grassin, Q., Manuguerra, J.-C. & Leclercq, I. Heat inactivation of the severe acute respiratory syndrome coronavirus 2. J. Biosaf. Biosecur. 3, 1–3 (2021).
Broughton, J. P. et al. CRISPR–Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 38, 870–874 (2020).
Joung, J. et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N. Engl. J. Med. 383, 1492–1494 (2020).
Patchsung, M. et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-020-00603-x (2020).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Knott, G. J. et al. Broad-spectrum enzymatic inhibition of CRISPR–Cas12a. Nat. Struct. Mol. Biol. 26, 315–321 (2019).
Smith, K. et al. CIDRE: an illumination-correction method for optical microscopy. Nat. Methods 12, 404–406 (2015).
Acknowledgements
We thank all members of the Hsu, Savage, Fletcher and Doudna laboratories for support and advice, and A. J. Ehrenberg, E. Charles and B. Thornton for helpful discussions. We thank M. Ott and R. Kumar for their help in coordinating the collection of natural saliva samples. This work was supported by the Shurl & Kay Curci Foundation, anonymous donors, Emergent Ventures, the National Institutes of Health (NIH) and DARPA under award N66001-20-2-4033. The views, opinions and/or findings expressed are those of the authors and should not be interpreted as representing the official views or policies of the Department of Defense or the US Government. We thank the NIH for their support (P.D.H. R01GM131073, DP5OD021369 and D.F.S. R01GM127463). J.A.D. is an Investigator of the Howard Hughes Medical Institute (HHMI). A.F. was supported by an NSF Graduate Research Fellowship.
Author information
Authors and Affiliations
Consortia
Contributions
S.S.C., S.A., A.F., A.R.J designed and performed experiments, analysed the data and prepared the manuscript. P.D.H. conceived the project, designed experiments, provided overall supervision for this work and wrote the manuscript, with input from all authors. D.F.S. and J.A.D. provided experimental input, edited the manuscript and co-supervised this work. B.C. provided microfluidic expertise and edited the manuscript. A.M.E., H.T., R.M. and M.X.T. designed and built the microfluidic cartridge and diagnostic instrument. S.S., A.B., N.A.S., M.A., A.R.H. and M.D.d.L.D. designed and built the optical instrument integrated with the microfluidic device, under the supervision of D.A.F. N.P. edited the manuscript and, with M.L., contributed to experimental design. Q.D. performed standard quantification experiments. D.C.J.S., T.Y.L. and G.J.K. purified proteins for assays and, with E.D. and S.K., provided biochemical expertise. A.M. developed computational methods for guide RNA selection with L.F.L., and S.B.B. and E.V.D. performed BSL-3 work with supervision from E.H. and S.A.S. The IGI Testing Consortium (J.R.H., E.L.-S., E.C.S., C.A.T., P.G., M.M., S.M., A.Z., I.S., A.C., M.C., and C.W.) performed diagnostic qPCR testing of the respiratory swab samples. E.A.M., K.K. and B.E. coordinated the collection of the saliva samples in the Salinas Valley farmworker study.
Corresponding authors
Ethics declarations
Competing interests
The Regents of the University of California and Wainamics have filed patents related to this work. P.D.H. is a co-founder of Spotlight Therapeutics and Moment Biosciences and serves on the board of directors and scientific advisory board, and is a scientific advisory board member to Vial Health and Serotiny. D.F.S. is a co-founder of Scribe Therapeutics and a scientific advisory board member of Scribe Therapeutics and Mammoth Biosciences. J.A.D. is a co-founder of Caribou Biosciences, Editas Medicine, Scribe Therapeutics and Mammoth Biosciences. J.A.D. is a scientific advisory board member of Caribou Biosciences, Intellia Therapeutics, eFFECTOR Therapeutics, Scribe Therapeutics, Mammoth Biosciences, Synthego, Algen Biotechnologies, Felix Biosciences and Inari. J.A.D. is a Director at Johnson & Johnson and has research projects sponsored by Biogen, Pfizer, AppleTree Partners and Roche. The other authors declare no competing interests.
Peer review
Peer review information
Nature Biomedical Engineering thanks Jin Wang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 rLAMP with T7-Floop primer mechanism.
FIP and BIP primers, assisted by F3 and B3 primers to XX, bind to the target DNA at F2c and B2c, adding complementary regions of DNA to the amplicon (F1c and B1c). Their complementarity to F1 and B1 result in loop structures, which facilitate further FIP and BIP binding at the loops (F2c and B2c) and extension, amplifying the target via formation of long concatemers. Floop and Bloop primers can additionally bind to the loops and extend, further increasing amplification. Addition of the T7 promoter at the 5’ end of the loop primer provides a substrate for T7 polymerase to transcribe, resulting in an RNA product containing inverted repeats of the target amplicon.
Extended Data Fig. 2 Validation of the microfluidic system.
(a) Screen of process control primer sets on commercial saliva samples. (b) Screen of T7 promoter locations on ACTB primer set on RNA extract and commercial saliva samples. (c) Schematic of cartridge design. The reaction can be separated in three steps: 1. amplification reaction; 2. post-amplification metering + Cas13 mix; 3. Cas13 reaction in detection chambers. Reagents stored on the cartridge are separated via a proprietary hydrophobic solution to avoid premature initiation of the reactions. After the LAMP reaction, the sample is split into two reactions: The left part of the cartridge will expose the sample to N gene crRNA (step 2a) while the right side of the cartridge will act as internal control with only ACTB crRNA (step 2b). (d) Results of on-board DISCoVER on 25 individual saliva samples (20 samples with SARS-CoV-2 genomic RNA at 500 cp/μL, 5 expected negative samples). A 2-fold fluorescence increase over blank cartridges at 10 minutes was the experimentally determined criteria for positive results. Samples with value below this threshold at 10 minutes were declared negative.
Supplementary information
Supplementary Information
Supplementary tables, figures and video caption.
Table
Sequences for LAMP, rLAMP, RT–qPCR and Cas13.
Video
Demonstration of the DISCoVER workflow.
Code
MATLAB script for the control of the imaging hardware.
Source data
Source Data Figs. 5c,d and 7d
Source data for Figs. 5c,d and 7d.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chandrasekaran, S.S., Agrawal, S., Fanton, A. et al. Rapid detection of SARS-CoV-2 RNA in saliva via Cas13. Nat. Biomed. Eng 6, 944–956 (2022). https://doi.org/10.1038/s41551-022-00917-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41551-022-00917-y
This article is cited by
-
Long-term monitoring of ultratrace nucleic acids using tetrahedral nanostructure-based NgAgo on wearable microneedles
Nature Communications (2024)
-
Fast and sensitive CRISPR detection by minimized interference of target amplification
Nature Chemical Biology (2024)
-
Biporous silica nanostructure-induced nanovortex in microfluidics for nucleic acid enrichment, isolation, and PCR-free detection
Nature Communications (2024)
-
Ultrasensitive single-step CRISPR detection of monkeypox virus in minutes with a vest-pocket diagnostic device
Nature Communications (2024)
-
Saliva-based detection of SARS-CoV-2: a bibliometric analysis of global research
Molecular and Cellular Biochemistry (2024)